Pyrrhic suppression of chemoreceptor signaling defects by Peter Ames and Sandy Parkinson |
|
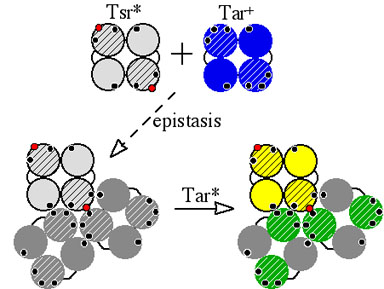
| Bacterial chemoreceptors of the methyl-accepting chemotaxis protein (MCP) family form supramolecular clusters with neural network properties. As described in a related poster, we found that certain tryptophan replacement mutations in Tsr, the serine chemoreceptor, not only abrogated Tsr function, but also blocked signaling by the wild-type aspartate chemoreceptor,Tar. We proposed that this epistatic or "jamming" effect involved physical interactions between Tsr and Tar molecules in mixed trimers of dimers (Fig. 1). To explore the nature of these receptor signaling teams, we attempted to isolate Tar mutants that were "resistant" to Tsr* jamming by selecting mutations in Tar that restored chemotactic ability to cells with an epistatic Tsr* defect. To our surprise, we obtained Tar mutations that suppressed the Tsr* signaling defect! We dubbed these mutations "pyrrhic suppressors" because they invariably destroyed Tar function (Fig. 1). | |
Figure 1. Proposed mechanism of pyrrhic suppression. The signaling domains of receptor dimers are shown schematically in cross-section with small circles representing wild-type (black) and mutant (red) trimer contact sites. Molecules in gray are nonfunctional. Epistatic Tsr mutants block Tar function by spoiling the conformation of mixed trimers of dimers (receptor signaling teams). Suppressor mutations in Tar (green) may produce an opposing conformational change in the receptor team that restores Tsr function but destroys Tar function. |
 |
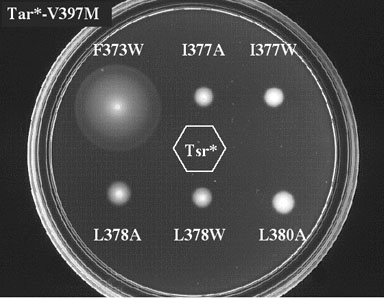
| According to the signal team model, pyrrhic suppression might occur through compensatory conformational interactions between Tar* and Tsr* molecules in a mixed trimer of dimers (Fig. 1). Conformational suppression of this sort should be highly allele- specific. Tests of Tar* suppressor mutations against a battery of Tsr* mutants with different trimer contact lesions confirmed the allele-specificity of the effect (Fig. 2). A particular Tar* suppressor only corrected a particular Tsr* defect, consistent with a structurally specific "lock and key" interaction between the Tar* and Tsr* molecules. | |
Figure 2. Allele-specificity of pyrrhic suppression. Shown are six diffferent epistatic mutants of Tsr carrying the same suppressor mutation in Tar. Only the Tsr- F373W mutant (which originally gave rise to the Tar-V397M mutation) regains chemotactic ability when combined with this Tar* suppressor mutation. Allele-specific suppression implies that the correction mechanism is conformational, involving direct interaction of the Tar* and Tsr* proteins. |
 |
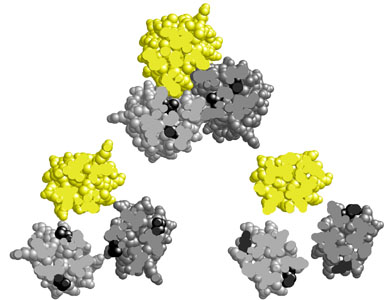
| The locations of the pyrrhic suppressor mutations in the Tar structure were also consistent with the proposed mechanism of conformational suppression (Fig. 3). Suppressor residues were often close to, but not at, the trimer contact sites. However, some suppressor mutations were located distal to the trimer interface, so more than one suppression mechanism could be at work. | |
Figure 3. Locations of pyrrhic suppressors in receptor signaling teams. Trimers containing one Tsr dimer (yellow) and two Tar dimers (gray) are shown in cross- section. All four Tar subunits within a trimer carry the same suppressor mutation, but each trimer illustrates a different suppressor site. In two of the trimers, one suppressor residue in each tar subunit appears to impinge directly on the trimer interface, which might account for the suppression effect. In the third trimer (lower right), none of the suppressor sites directly interacts with the trimer interface. This suppressor may perturb trimer geometry by a different mechanism. |
 |
| Pyrrhic suppression provides a powerful experimental tool for investigating the functional architecture of chemoreceptor signaling teams. Because neither Tar* nor Tsr* molecules alone are functional, we can mix them at different stoichiometries, either in vivo or in vitro, to manipulate the composition of active signaling teams. Stay tuned for further developments . . . | |
| Ames, P., C.A. Studdert, R.H. Reiser, and J.S. Parkinson (2002). Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. | |
| Ames, P. and J.S. Parkinson (2006). Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. USA 102:9292-9297. | |
